![]() Go to frame view (Recommended only for
screen resolution 1024x768)
Go to frame view (Recommended only for
screen resolution 1024x768)
10 Catalytic Activity of Nucleic Acids
The totally unexpected discovery of the catalytic activity of RNAs was one of the highlights of molecular biology in the eighties, has turned out to be a rather common phenomenon. The RNA enzymes isolated from different sources have become known as ribozymes.
Ribozymes catalyze reactions of specific cleavage or exchange of phosphodiester bonds in polyribonucleotides and in some instances also in polydeoxyribonucleotides. They catalyze transfer of the phosphate group and nucleotides; in the latter case, the processes may include both specific cleavage of the polynucleotide chain and its extension. As of today, the substrates of ribozymes in the overwhelming majority of cases are nucleic acids themselves. In other words, the organization of active centers of ribozymes and binding of substrates by them are due to specific RNA-RNA or RNA-DNA interactions. Ribozymes satisfy all the criteria met by enzyme proteins: the reactions they catalyze are highly specific, and the rate of these reactions is increased by them as much as 1011-fold with their initial structure remaining virtually intact.
10.1 RNA subunit of Ribonuclease P
The first and only catalytic RNA known to act as an enzyme in vivo was discovered in 1983 by S. Altman, N. Pace and co-workers. It had turned out to be an RNA subunit of ribonuclease P - the key enzyme in the processing of tRNAs, instrumental in the maturation of their 5' ends (Fig. 10-1A). At first it was found that RNase P is a ribonucleoprotein containing, along with a protein (ca. 14 kD), a small RNA subunit (ca. 400 nucleotides). It is only the latter that displayed catalytic activity. In contrast to the complete enzyme, this activity manifested itself only at high concentrations of magnesium ions which, on the one hand, were conducive to compact folding of the RNA structure and, on the other, screened the negative charges of the phosphate groups in the enzyme and substrate and participated directly in the chemical step of the cleavage reaction. The ribozyme-substrate interactions were characterized by a value of Km close to that of the holoenzyme, the ribozyme cleaving the pre-tRNA at a rate only half as high as RNase P itself. Just as the complete enzyme, ribozyme cleaves the pre-tRNA in such a way that the phosphate group remains at the 5' end of the tRNA.
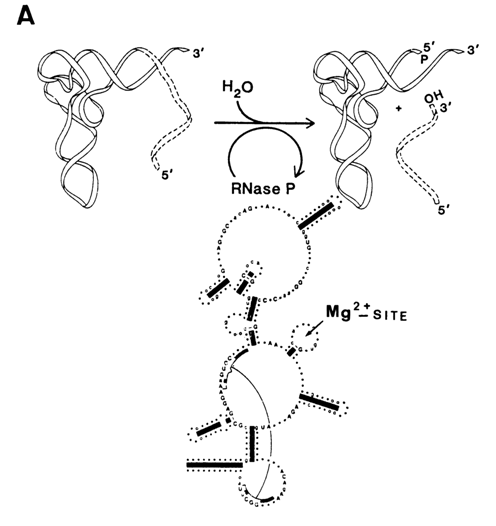
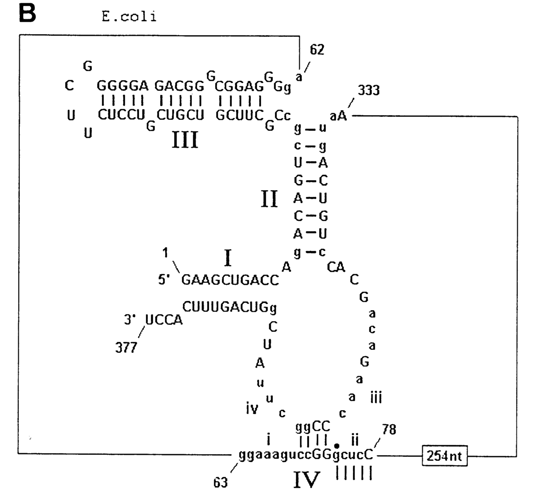
Fig. 10-1. Ribonuclease P. A. Scheme illustrating participation of the catalytic RNA subunit of RNase P in tRNA maturation. The consensus secondary structure of eubacterial RNA is shown. Filled rectangles represent phylogenetically proven helices. Invariant nucleotides are indicated by upper letters. The shaded areas include RNA sequences which are essential for catalysis (adapted from 3. W Brown and N. Pace, Nucleic Acids Res., 20, 1451-1456 (1992). B and C. Hypothetical cage structures for RNA subunits of RNase P of E. coli (B) and B. subtilis (C). Nucleotides depicted in lowercase letter are concerved. Nucleotides 74-78 in B and 57-60 in C form base pairs with regions in the central portions of the RNA (adapted from A. C. Forster and S. Altman, Cell, 62, 407-409 (1990)).
The secondary structure of the RNA subunit of RNase P was established by phylogenetic analysis. Figures 10-1 B and C illustrate the arrangement of polynucleotide chains in those portions of the RNA subunits of the enzymes isolated from E. coli and B. megaterium which, judging from the results of phylogenetic analysis, are especially important for the enzymatic activity of ribozymes. Their structures were given in their entirety in Chapter 8 as an illustration of comparative (phylogenetic) analysis in selecting the secondary RNA structure model (Fig. 8-40). Interestingly, when applied to RNase, such analysis also revealed that the helical elements present only in E. coli RNA and only in B. megaterium RNA are not essential insofar as the enzymatic activity of ribozymes is concerned. This finding has made it possible to derive, from a gene of the RNA subunit of E. coli RNase P, a so-called miniribozyme 263 nucleotides long, which did not differ much from the initial ribozyme in terms of catalytic properties. Nevertheless, attempts to identify the real catalytic center of the RNA subunit of RNase P are yet to be successful. The only thing that has become clear is that these ribozymes recognize in their substrates a certain motif in the three-dimensional structure of pretRNAs, rather than their primary structure, because the sites of pre-tRNA cleavage by the ribozyme differ widely in nucleotide sequence.