![]() Go to frame view (Recommended only for
screen resolution 1024x768)
Go to frame view (Recommended only for
screen resolution 1024x768)
1.9 Pseudouridine
Apart from major and minor nucleosides with N-glycosidic bonds some RNAs also contain sizable amounts of an unusual nucleoside with a different type of glycosidic bond, known as pseudouridine. This unusual, or "fifth", nucleoside was discovered as far back as 1951, but it took ten years to decipher its structure in spite of the fact that the chief methods for establishing the structure of nucleosides had been developed by the early fifties and become available to many laboratories around the world. The conventional methods for determining a structure did not work on pseudouridine. It was soon found out that the unusual nucleoside has the same chemical composition as uridine. However, unlike uridine, it is extremely stable under conditions of acid hydrolysis, which explains why attempts to break it down into components failed for quite some time. The UV absorption spectrum of the compound differed from that of uridine and 1-alkyluracils and was closely similar to that of 5-hydroxymethyluracil. These findings had led to a conclusion that the "fifth nucleoside" is a derivative of uracil, yet, in contrast to the latter, represents a C-glycoside with glycosidic bonding at C5 or C6 in the pyrimidine ring. This conclusion was supported by the results of UV spectroscopy at different values of pH.
The bathochromic shift of the absorption curve corresponding to the nucleoside of interest in the UV region at pH > 8.0, as compared to a more acid medium, suggested that the heterocyclic base of this nucleoside contains two hydroxy groups prone to ionization.
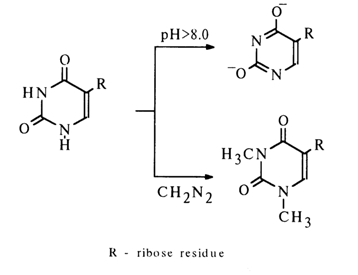
Methylation of both nitrogens of the base was significant in demonstrating that they are not substituted in the starting nucleoside. The presence of D-ribose was proved by hydrazine-induced cleavage of the nucleoside. Thus, it was established that the "fifth nucleoside" differs from uridine only by the nature of its glycosidic bond. It is for this reason that the nucleoside was called pseudouridine.
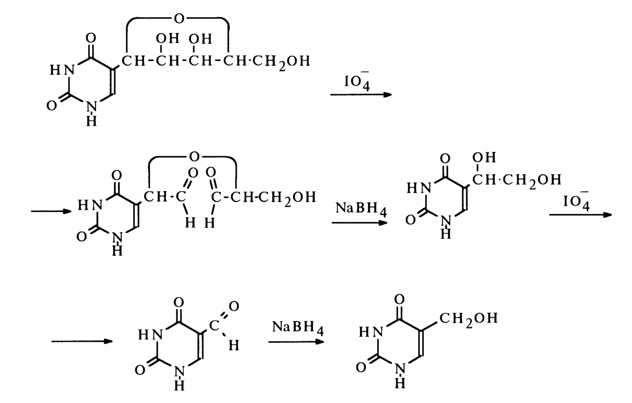
The fact that pseudouridine has the structure of 5-ribofuranosyluracil was established definitively through double oxidation with periodic acid followed by reduction with sodium borohydride - that is, through conversion into 5-hydroxymethyluracil.
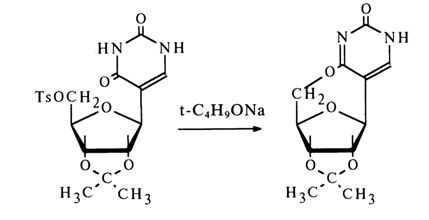
The results of NMR spectroscopy, attesting to presence of a proton at C6 in pseudouridine and its absence at C5, corroborated the findings of the preceding studies. The structure of pseudouridine, just as that of 5-D-ribofuranosyluracil, was also confirmed by synthesis. Ale configuration of the glycoside center was established in the usual manner:
Thus, pseudouridine is a 5-P-D-ribofuranosyluracil. Given below are the commonly recognized structural formula and abbreviated symbols of this nucleoside:
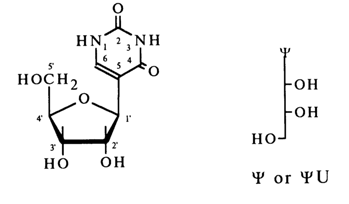
Some RNAs have also been found to contain 2'-O-methylpseudouridine (designated by the abbreviated symbol Wm).